We are fascinated by the fundamental question of how the internal organization of cells is established, maintained and regulated to generate the functional polarity seen in eukaryotic cells. Just imagine what would happen if cells did not have the ability to polarize: they wouldn’t know which axis to divide along, epithelial cells wouldn’t be able to transport, secretory cells would secrete things to the wrong surface, cells wouldn’t be able to migrate or chemotax, cells wouldn’t be able to undergo asymmetric divisions that generate cellular diversity during development, etc.! Life would not be possible!!
********
We study two model systems to delve into a molecular understanding of polarity:
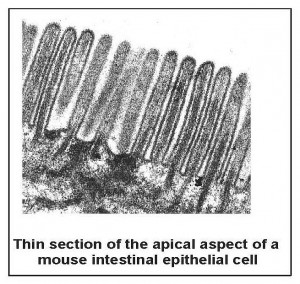
The first is to elucidate the structure and function of the apical aspect of polarized epithelial cells (Figure 1). These cells have distinct apical and basolateral domains that allow them to perform their transport function. Establishment of polarity is directed by cues supplied by the extracellular matrix and adjacent cells, which the cell interprets using signal transduction pathways to assemble a polarized cytoskeleton.
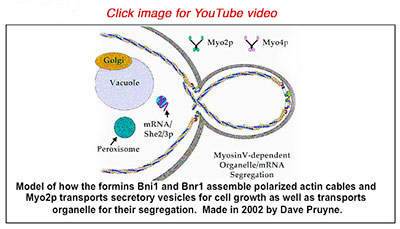
The second system utilized is the budding yeast Saccharomyces cerevisiae. We are using this system to determine the mechanism the cell uses to undergo polarized growth and segregate its organelles during the cell cycle. During cell growth in yeast, the mother cell picks a region on the cell surface to which secretion is targeted for the assembly of a bud, and then all organelles have to be segregated along this axis during cell division. Below is short video of one division cycle in the budding yeast:
Investigating both these systems has lead us to study not only the components of microfilaments, actin and actin-binding proteins, but also their regulation by signal transduction pathways and how they participate in membrane traffic pathways.
Our studies use diverse biochemical, structural, molecular genetic and cell biological approaches.
*********
Our Two Major Projects Are:
1. An investigation into the molecular organization, regulation and function of the microfilaments that make up the microvilli on the apical aspect of polarized cells.
(a) Function and regulation of the ERM Proteins and Merlin:
We are studying all the components of the apical cortical cytoskeleton of epithelial cells. We have identified and characterized several microfilament-associated proteins (including villin, fimbrin, brush border myosin I, and ezrin) and, together with ultrastructural studies, have begun to piece together the functions of each component (reviewed in 1-4). Our current focus is on ezrin, a protein we discovered enriched in cell surface microvilli (5) (Figure 2A and 2B), that we named in honor of Ezra Cornell. (Click onto any photo for larger resolution.)
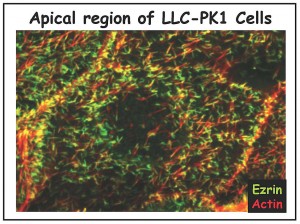
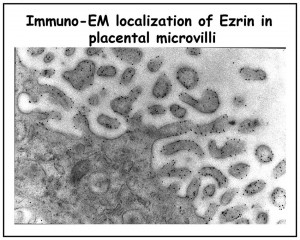
Ezrin is the founding member of the ERM (ezrin-radixin-moesin) protein family, closely related to the tumor suppressor merlin and a member of the Band 4.1 superfamily (Figure 3). The established function of ezrin is to provide a regulated linkage between the actin cytoskeleton and the plasma membrane in specific cell surface structures.
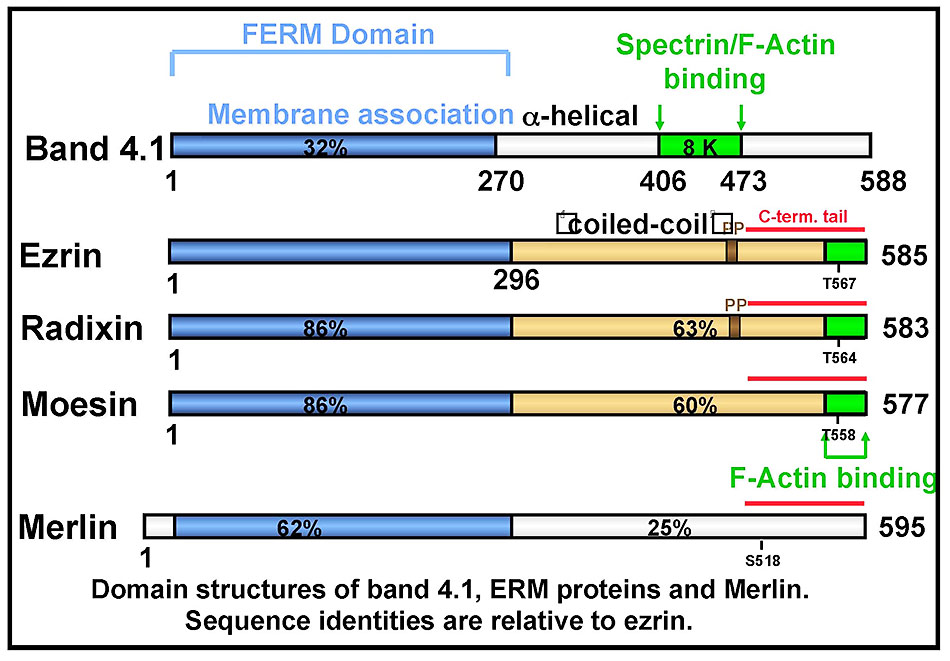
Our early work showed that ezrin (and radixin and moesin) is regulated by conformational masking (Figure 4): in the dormant form, a high affinity intra-molecular association exists between the FERM domain and the C-terminal tail (6). Dissociation of this intramolecular interaction is necessary to unmask binding sites for several ligands of ezrin, including the scaffolding protein EBP50 (7,8) and F-actin (6). Work from a number of labs has indicated that ezrin activation is regulated by signaling pathways, most likely by binding PI(4,5P)2 and subsequent phosphorylation on threonine 567 (4).

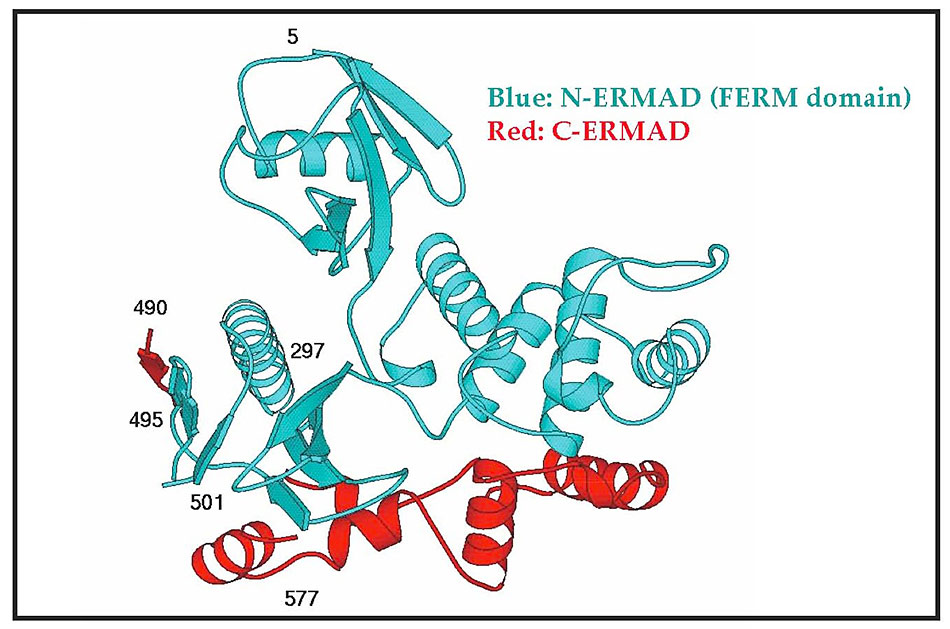
Since ezrin undergoes conformational regulation and is implicated in multiple protein-protein interactions (see below), it is important to understand these conformational changes at the atomic level to reveal how binding sites become unmasked. Towards this goal, we have collaborated with Dr. P. Andrew Karplus (formerly a colleague at Cornell, now at Oregon State University, Corvallis) on the X-ray structure of various forms of ezrin alone and complexed with its ligands. So far, we have determined the structure of the associated FERM (blue) and C-terminal tail (red: ‘C-ERMAD’) of moesin (Figure 5), thus revealing the interface between these domains involved in masking binding protein interaction sites (9). We have also determined the structural basis of masking of the EBP50 binding site in dormant ezrin by determining the surface on the FERM domain to which EBP50 binds (10), and the structural changes that occur in the FERM domain upon release of the C-terminal tail (11).
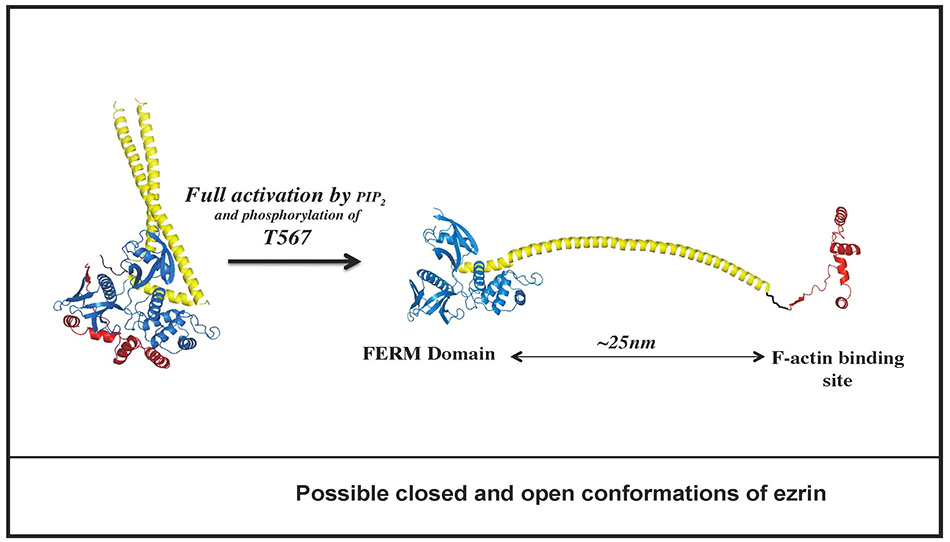
In collaborative work with the Tesmer lab at the University of Michigan, we have reported on the structure of full length dormant ERM protein from insect cells (12) (Figure 6A and video, below). As you can see, the central α-helical domain forms an anti-parallel coiled-coil structure. (Click on photos for larger resolution.)

Based on biophysical studies of wild type and mutant proteins, we believe that the molecule is spring-loaded, so that upon activation the C-terminal domain (red) dissociates from the FERM (blue) allowing the central domain to extend into a long α-helix spanning about 25nm between the membrane associated (FERM) domain and the F-actin binding site (located in the last 30 residues of the C-terminal tail) (Figure 6B).
Although several ligands have been identified that bind ezrin, we are currently using proteomic approaches to identify proteins that selectively bind active, but not inactive, ezrin as these are likely to be the most relevant physiologically. We have made use of SILAC (Stable Isotope Labeling by Amino acids in Culture) combined with mass spectrometry to specifically identify proteins that associate with wild type ezrin (that can be activated) versus ezrin-T567A (that cannot be activated). This has revealed a number of new microvillar proteins. Among these are the kinases LOK and SLK, which we have found are localized preferentially to the apical membrane of epithelial cells. Moreover, LOK and SLK are the major kinases necessary for ezrin’s activation by threonine-567 phosphorylation and required for the presence of microvilli on epithelial cells. Additionally, we have found that this threonine-567 phosphorylation is dynamic in the sense that phospho-ezrin turns over every minute or so. Stabilizing the phosphorylated form leads to mis-localization of ezrin basolaterally. Thus, dynamic cycling of ezrin threonine-567 phosphorylation driven by LOK/SLK is necessary for restricting ezrin function to the apical domain (13).
We have also studied merlin, the product of the neurofibromatosis II tumor suppressor gene as it is closely related to ezrin (see Figure 3). Many studies show that merlin, like ezrin, is conformationally regulated through association between its FERM domain and C-terminal tail. Moreover, ezrin and merlin bind many of the same ligands, and the two proteins can form heterodimers (14). However, since merlin and ERM proteins perform distinct essential functions in both vertebrate and lower organisms (e.g. the fly, summarized in ref. 3, 4), we are trying to uncover the related and distinct contributions of merlin and ERM proteins to cellular organization and function. One surprising difference between merlin and ERM proteins is a model that suggests active merlin is in a closed state that is inactivated by phosphorylation on serine 518 to open up the molecule (summarized in 3). This model is the exact opposite from ezrin in which the open form is active; we have shown that this model for merlin is not correct. Like ezrin, the open state is active, but in merlin’s case, phosphorylation enhances the closed inactive state (15).
(b) Function and regulation of the scaffolding protein EBP50.
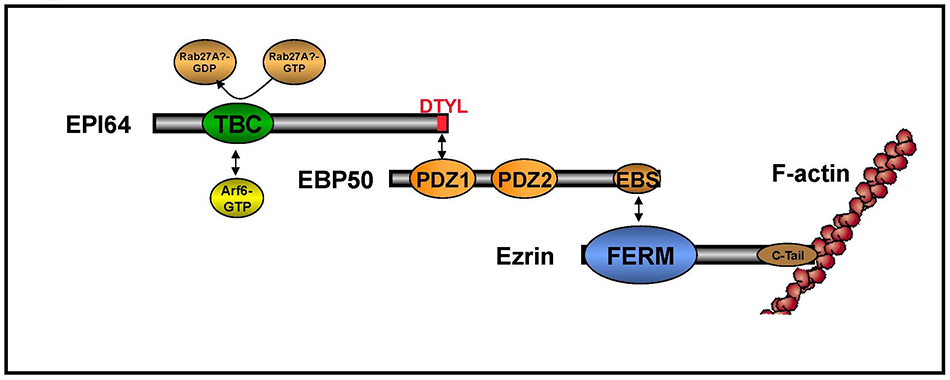
As discussed above, ezrin has to be activated to bind its ligands. Activated ezrin can bind directly to specific membrane proteins through its FERM domain, and indirectly through the scaffolding protein EBP50. EBP50 (also known as NHERF1) has two PDZ domains (PDZ1 and PDZ2) and a C-terminal region that binds with very high affinity to the FERM domain of ezrin (7, and see Figure 4). Thus, EBP50 can link the cytoplasmic domains of specific membrane proteins to ezrin and F-actin to regulate their function and/or location (2-4).
Recent work has shown that EBP50 is necessary for the assembly of microvilli on cultured epithelial cells, and that this requires functional PDZ1 and ezrin-binding domains. Interestingly, when phosphorylated by PKC or Cdc2, EBP50’s function in microvillar biogenesis is lost. Biochemical analysis of EBP50 mutants shows that in phosphorylated EBP50, ligand binding to PDZ2 results in a conformational change to render PDZ1 inaccessible to ligands, and therefore become non-functional in vivo (16). This study also documented that EBP50 is an elongated monomer, rather than forming oligomers as suggested by other laboratories.
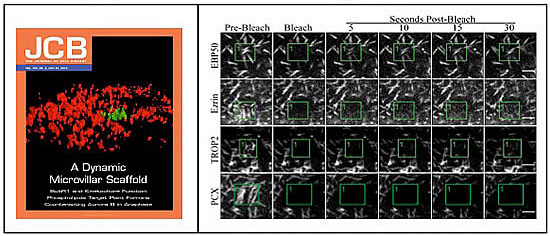
Microvilli are not static structures (see video, below, of GFP-EBP50), so we are examining the dynamics of both microvilli and the individual components using fluorescence bleaching techniques in living cells. Remarkably for a scaffolding protein, we have recently found that EBP50 is much more dynamic than ezrin in vivo (Figure 7A, B), and this dynamics is reduced when either PDZ domain of EBP50 is mutated to be non-functional (17).
We have also studied another scaffolding protein in the EBP50 family, PDZK1, that has four PDZ domains. It is also found in the apical domain, but does not bind ezrin. Remarkably, the C-terminal tail of PDZK1 binds to PDZ1 of EBP50, thereby extending the scaffolding function of EBP50 (18, 19) (Figure 4). As also shown in Figure 4, ezrin, EBP50 and PDZK1 all undergo intramolecular interactions that have to be opened in order for the proteins to function.
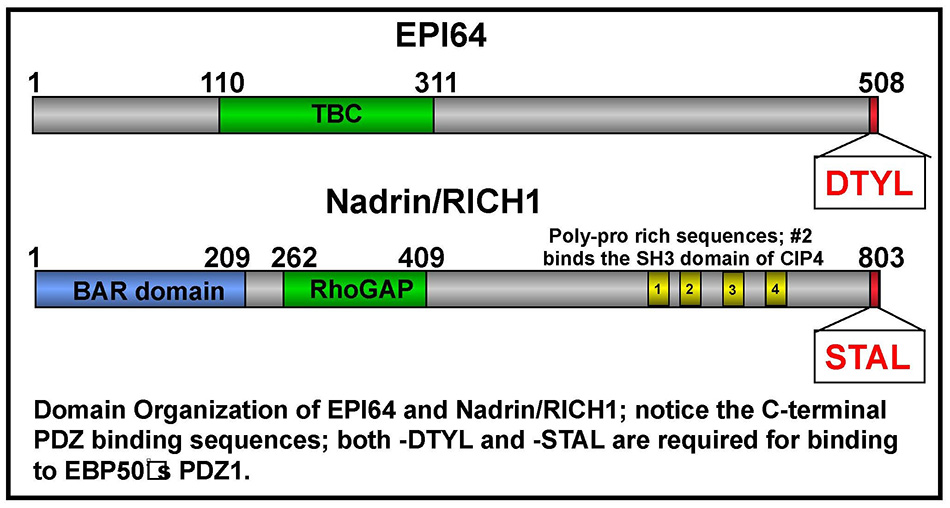 In earlier work, we set out to identify membrane proteins that bind the PDZ domains of EBP50. We were surprised to find that the first PDZ domain of EBP50 binds the C-terminal tail of some cytoplasmic proteins, including EPI64 and nadrin (20) (Figure 8A). EPI64 has a TBC domain that has both a RabGAP activity and binds Arf6-GTP (Figure 8B) to regulate both membrane traffic and the presence of microvilli on the surface of epithelial cells (21). Recently, we have shown that EPI64 regulates a Rab8 – dependent pathway, but how this contributes to the presence of microvilli has not yet been elucidated (22). Nadrin is a BAR- and RhoGAP-containing protein that associates with EBP50, although the function of this interaction is not yet clear. (Click on photos for larger resolution.)
In earlier work, we set out to identify membrane proteins that bind the PDZ domains of EBP50. We were surprised to find that the first PDZ domain of EBP50 binds the C-terminal tail of some cytoplasmic proteins, including EPI64 and nadrin (20) (Figure 8A). EPI64 has a TBC domain that has both a RabGAP activity and binds Arf6-GTP (Figure 8B) to regulate both membrane traffic and the presence of microvilli on the surface of epithelial cells (21). Recently, we have shown that EPI64 regulates a Rab8 – dependent pathway, but how this contributes to the presence of microvilli has not yet been elucidated (22). Nadrin is a BAR- and RhoGAP-containing protein that associates with EBP50, although the function of this interaction is not yet clear. (Click on photos for larger resolution.)
Overall, our studies on the apical aspect of epithelial cells have provided insight into how microvilli are regulated and assembled, and the contribution this has on the functional polarity of these cells.
2. The mechanism of polarized growth and organelle segregation during the cell cycle in the budding yeast Saccharomyces cerevisiae.
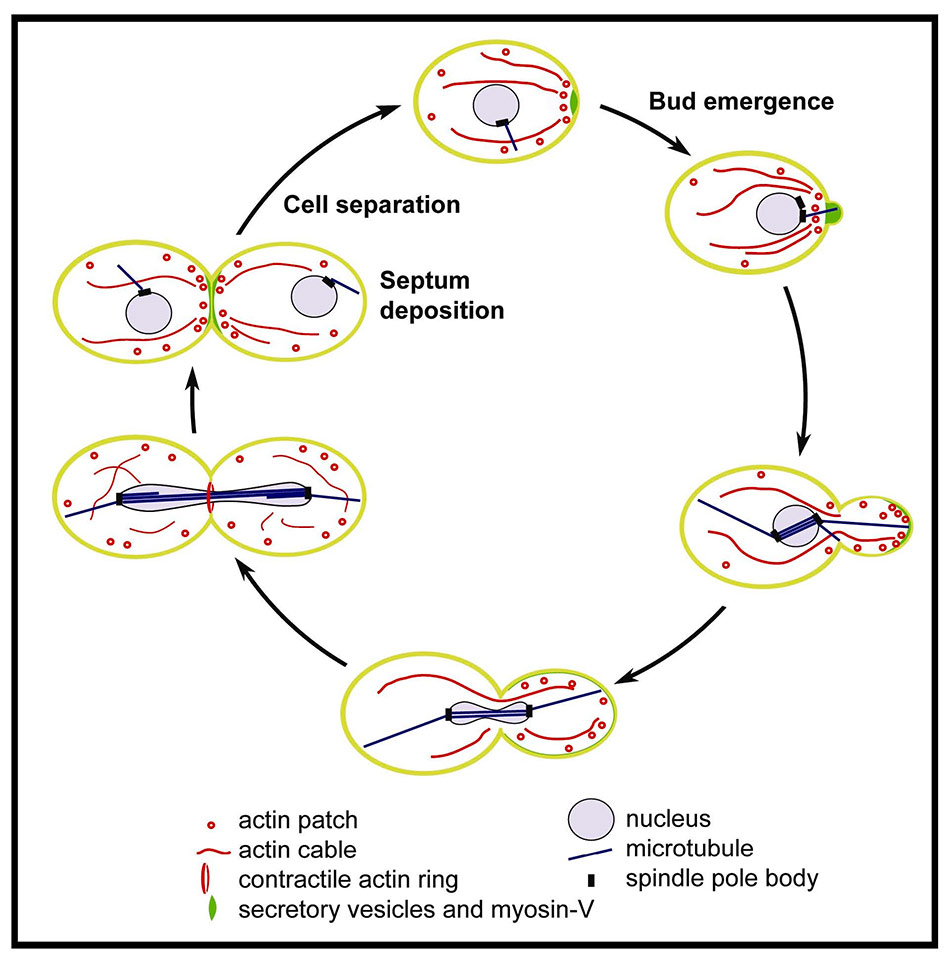
To grow, yeast has to target secretion to the growing bud. This polarized growth is a result of vectorial delivery of post-Golgi secretory vesicles (see video, below), which are delivered and targeted by a polarized actin cytoskeleton. The yeast actin cytoskeleton exists in three distinct types of structures: cables that extend through the cell to sites of polarized growth, cortical patches that also cluster at sites of polarized growth, and the contractile ring at cytokinesis (see Figure 9; reviewed in 23, 24).
Work from others has shown that cortical patches are involved in endocytosis, whereas we focus on the structure and function of the actin cables. We and others have shown that actin cables are involved in directing polarized growth and segregation of organelles during the cell cycle. Our work therefore has focused on: (a) the assembly and dynamics of actin cables, and (b) the myosin-V motor that transports cargo along them.
(a) How are actin cables assembled?
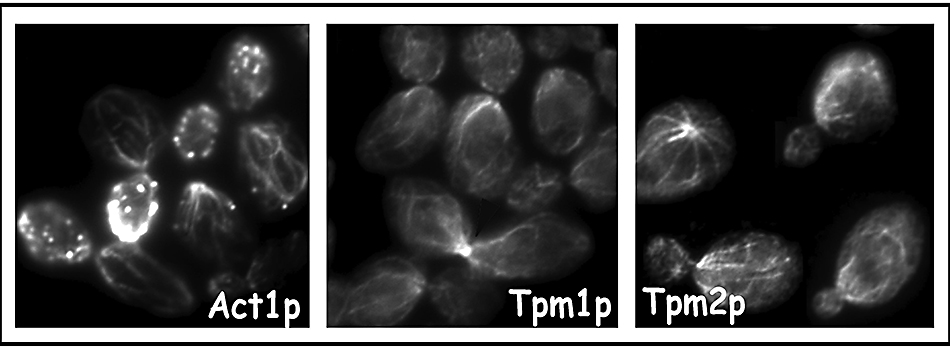
A critical early finding was that yeast tropomyosins (encoded by theTPM1 and TPM2 genes specifying two related isoforms), are a specific component of actin cables and provide an essential function (25, 26) (Figure 10). Using a conditional tropomyosin mutant (tpm2Δ tpm1-2), we found that tropomyosin is necessary to stabilize actin cables, and they, not cortical patches, determine the polarity of secretion (27).
Since polarized actin cables are the tracks for Myo2p-dependent organelle transport (see below), a key question is how cables are established, maintained and regulated. In collaboration with Marie Evangelista and Charlie Boone at the University of Toronto, we were able to show that the formins Bni1p and Bnr1p together provide an essential Arp2/3-independent function in the assembly of actin cables (28).
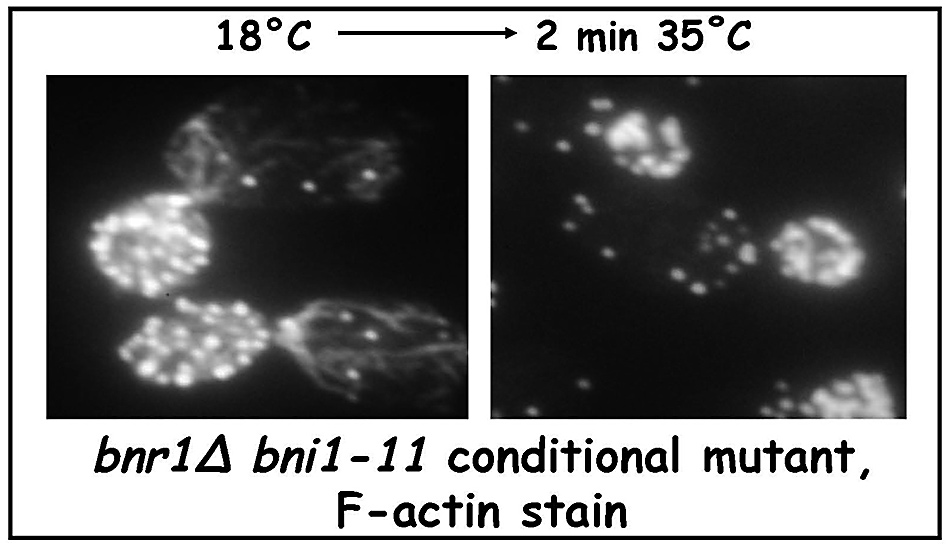
Thus a conditional bnr1Δ bni1-11 strain has actin cables at the permissive temperature, but the cables disappear after a quick shift to the restrictive temperature (Figure 11).
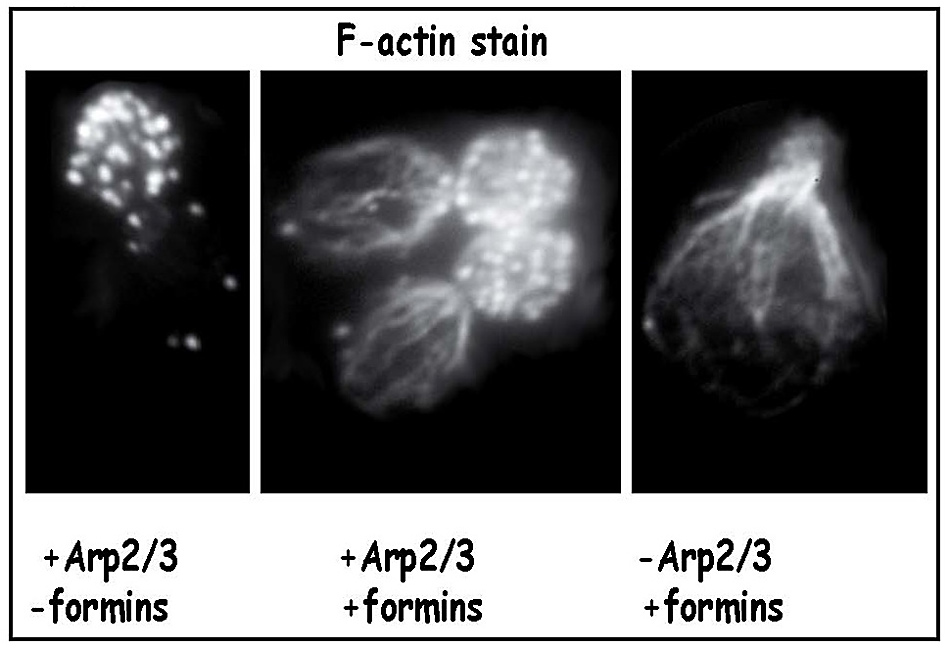
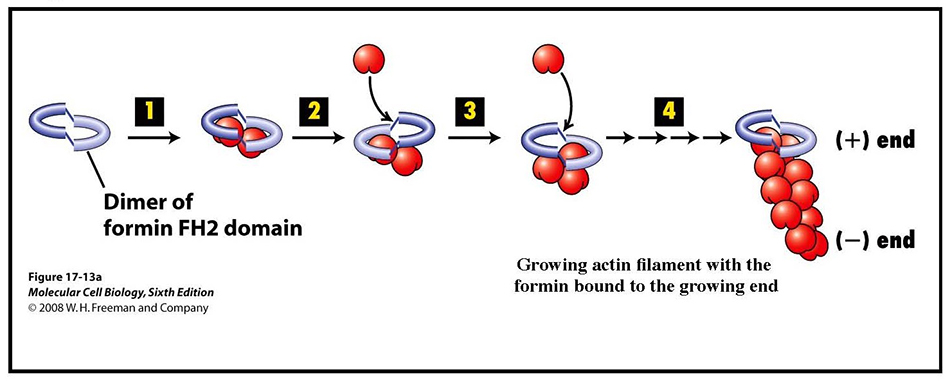
Since cells lacking Arp2/3 lack cortical patches yet have cables, and cells lacking formin function have patches but not cables, patches and cables are assembled by different mechanisms (Figure 12). Our subsequent collaborative work, with additional help from Sally Zigmond at the University of Pennsylvania, showed that formins are able to nucleate the assembly of actin filaments. Specifically, the adjacent FH1+FH2 domains of formins collaborate to nucleate the assembly of actin filaments and then remain bound to the plus, or barbed, end of the filament as it continues to elongate (29) (shown schematically in Figure 13).
Subsequent studies have been aimed at understanding the regulation of yeast formins (30) and the distinct contributions of yeast’s two formins, Bni1p and Bnr1p (31), and how yeast distributes actin between different organizations (32). Surprisingly, both Bnr1p and Bni1p have multiple regions that each can localize the protein to correct sites for actin assembly (33, 34).
(b) What motor transports secretory vesicles and other organelles down actin cables?
Actin cables are oriented with the plus end at sites of cell growth. Since myosin motors in general move towards the plus end of actin filaments, we looked for yeast myosins that could transport secretory vesicles. Our early work showed that the unconventional myosin V encoded by the MYO2 gene is the molecular motor that delivers secretory vesicles down actin cables to their site of exocytosis (27, 35). Remarkably, this same myosin is responsible for orienting the nucleus in order to align the mitotic spindle with the axis of cell division. In collaboration with Tim Huffaker’s lab in our Department, we have shown it does this by binding through its tail to a protein called Kar9p, which itself binds to Bim1p bound to the end of cytoplasmic microtubules (37).
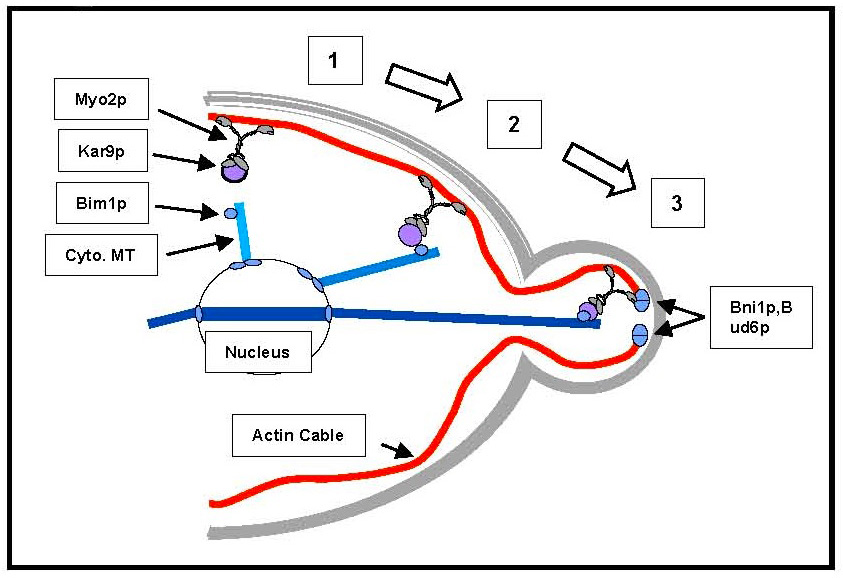
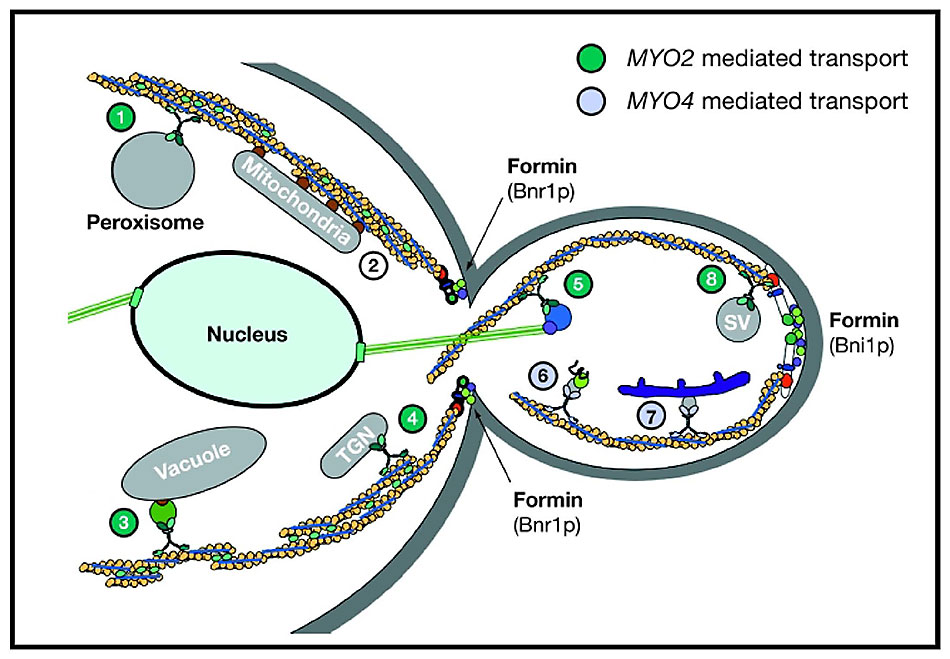
Myo2p is also responsible for segregation of the vacuole, peroxisomes and the trans-Golgi network (Figure 15). Myo2p has to bind to each of these cargos through cargo-specific receptors and transport the organelles at the right time during the cell cycle. We are investigating the identity of cargo receptors and the delivery cycle of Myo2p.
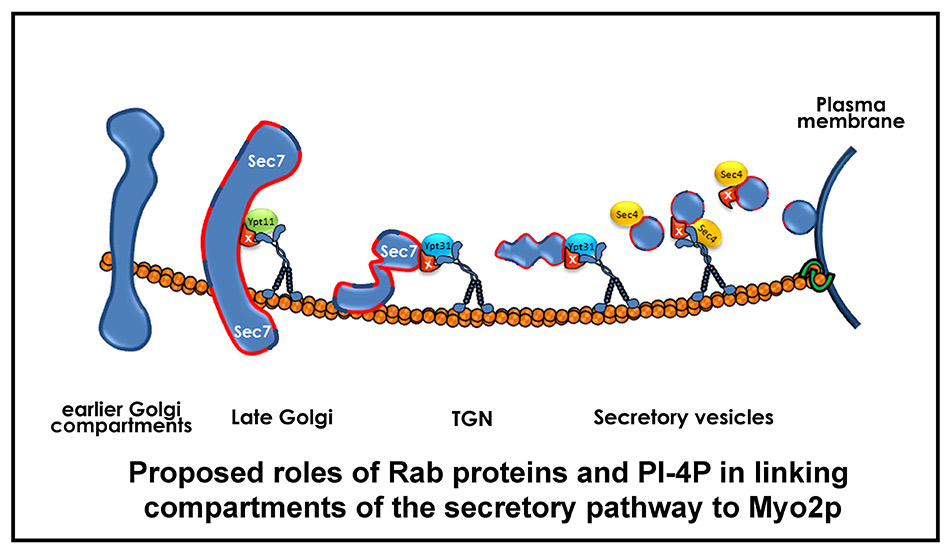
Recent work has provided insight into how Myo2p transports compartments of the secretory pathway. Myo2p binds directly to the Rab proteins Ypt31/32p (the yeast Rab11 homologue) associated with the TGN, and to Sec4p (the Rab8 homologue) associated with post-Golgi secretory vesicles. Both these Rabs are necessary for Myo2p to bind and transport these compartments to sites of cell growth. In addition, we have found that Golgi-synthesized PI4P is also necessary for Myo2p to bind these cargos, and the cell uses a coincidence detection mechanism involving a Rab protein and PI4P to bind to and transport these compartments (38, 39; Figure 16). Genetic evidence suggests there are additional components of the secretory vesicle receptor; current studies are aimed at identifying them.
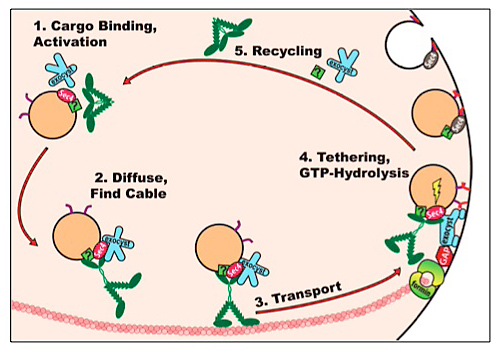
What is the delivery cycle of Myo2p in its vectorial transport of secretory vesicles? We have recently used high resolution imaging to define features of the cycle by which Myo2p delivers secretory vesicles (Figure 17). First, we have shown that Myo2p activation requires secretory cargo. Second, about 10 motors associate with each secretory vesicle, which is transported along actin cables to sites of cell growth. Third, upon arrival, motor release requires a functional exocyst – the tethering complex necessary for exocytosis. Interestingly, motor release does not need exocytic vesicle fusion. Since the exocyst and Myo2p are both effectors of Sec4p-GTP, this suggests that GTP hydrolysis may integrate exocytic function with motor release. Indeed, mutations that delay GTP hydrolysis affect both exocytosis and motor release (40).
In aggregate, our and other labs’ studies have provided a model (see video, below) of how yeast assembles polarized actin cables and uses them for the delivery of secretory vesicles and segregation of organelles (41).
**************************
References:
Apical Aspect of Epithelial Cells:
1. Bretscher, A. (1991). Microfilament structure and function in the cortical cytoskeleton. Ann. Rev. Cell Biol. 7, 337-374.
2. Bretscher, A., Chambers, D., Nguyen, R. & Reczek, D. (2000) ERM-merlin and EBP50 protein families in plasma membrane organization and function. Ann. Rev. Cell & Dev. Biol. 16, 113-143.
3. Bretscher, A., Edwards, K. & Fehon, R. (2002) ERM proteins and merlin: integrators at the cell cortex. Nature Reviews Molecular and Cell Biology 3, 586-599.
4. Fehon, R. G., McClatchey, A. I. & Bretscher, A. (2010). Organizing the Cell Cortex: The role of ERM proteins. Nature Reviews Molecular Cell Biology, 11, 276-287.
5. Bretscher, A. (1983). Purification of an 80,000 dalton protein that is a component of the isolated microvillus cytoskeleton, and its localization in non-muscle cells. J. Cell Biol. 97, 425-432.
6. Gary, R. & Bretscher, A. (1995). Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol. Biol. Cell 6, 1061-1075.
7. Reczek, D., Berryman, M. & Bretscher, A. (1997) Identification of EBP50: a PDZ domain containing phosphoprotein that associates with members of the ERM family. J. Cell Biol. 139, 169-179.
8. Reczek, D. & Bretscher, A. (1998). The carboxy-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant monomer. J. Biol. Chem. 273, 18378-18384.
9. Pearson, M., Reczek, D., Bretscher, A. & Karplus, P. A. (2000). Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin-binding tail domain Cell 101, 259-270.
10. Finnerty, C., Chambers, D., Ingraffea, J., Faber, H. R., Karplus, P. A. & Bretscher, A. (2004). The EBP50-moesin interaction: structural analysis of a binding site regulated by direct masking on the FERM domain. J. Cell Sci. 117, 1547-1552.
11. Smith, W.J., Nassar, N., Bretscher, A., Cerione, R. A. & Karplus, P.A. (2003). Structure of the active FERM Domain of Ezrin: conformational and mobility changes identify keystone interactions. J. Biol. Chem. 278, 4949-4956.
12. Li, Q., Nance, M. R., Kulikauskas, R., Nyberg, K., Fehon, R., P., Karplus, P. A., Bretscher, A. & Tesmer, J. J. G. (2006). Self-masking in an intact ERM-merlin protein: an active role for the central a-helical domain. J. Mol. Biol. 365,1446-59.
13. Viswanatha, R, Ohouo, P., Smolka, M. B. & Bretscher, A. (2012). Local cycles of LOK/SLK-dependent phosphorylation restruict ezrin function to the apical aspect of epithelial cells. J. Cell Biol. in press.
14. Nuygen, R., Reczek, D. & Bretscher, A. (2001). Heirarchy of N- and C-ERMAD associations and common ligands between ezrin and merlin. J. Biol Chem. 276, 7621-7629.
15. Sher, I., Hanemann, C. O., Karplus, P. A. & Bretscher, A. (2012). The tumor suppressor merlin controls growth in its open state and is converted by phosphorylation to a less-active more-closed state. Developmental Cell 22, 1-3.
16. Garbett, D., Lalonde, D. & Bretscher, A. (2010). The Scaffolding protein EBP50 regulates microvillar assembly in a phosphorylation-dependent manner. J. Cell Biol. 191, 397-413.
17. Garbett, D. & Bretscher, A. (2012). PDZ interactions regulate rapid turnover of the scaffolding protein EBP50 in microvilli. J. Cell Biol. 198, 195-203.
18. Lalonde, D. & Bretscher, A. (2009) The scaffold protein PDZK1 undergoes a head-to-tail intramolecular association that negatively regulates its interaction with EBP50. Biochemistry 48, 2261-2271.
19. Lalonde, D., Garbett, D. & Bretscher, A. (2010) A regulated complex of the scaffolding proteins PDZK1 and EBP50 with ezrin contribute to microvillar organization. Mol. Cell Biol. 21, 1519-1529.
20. Reczek, D. & Bretscher, A. (2001) Identification of EPI64, a TBC/rabGAP domain-containing microvillar protein that binds to the first PDZ domain of EBP50 and E3KARP. J. Cell Biol. 153, 191-206.
21. Hanono, A., Garbett, D., Reczek, D., Chambers, D. N. & Bretscher, A. (2006). EPI64 regulates microvillar sub-domains and structure. J. Cell Biol. 175, 803-813.
22. Hokanson, D. & Bretscher, A. (2012). EPI64 interacts with Slp1/JFC1 to coordinate Rab8a and Arf6 membrane trafficking. Mol. Biol. Cell, 23, 701-715.
Cell Polarity in Budding Yeast:
23. Schott, D., Huffaker, T. & Bretscher, A. (2002). Microfilaments and microtubules: the news from yeast. Curr. Opin. Microbiol. 5, 564-574.
24. Pruyne, D., Legesse-Miller, A., Gao, L., Dong, Y. & Bretscher, A. (2004). Mechanisms of polarized growth and organelle segregation in yeast. Ann. Rev. Cell Dev. Biol. 20, 559-591.
25. Liu, H. & Bretscher, A. (1989). Disruption of the single tropomyosin gene in yeast leads to a disappearance of actin cables from the cytoskeleton. Cell 57, 233-242.
26. Drees, B., Brown, S., Barrell, B. and Bretscher, A. (1995). Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J. Cell Biol. 128, 383-392.
27. Pruyne, D., Schott, D. & Bretscher, A. (1998). Tropomyosin-containing actin cables are the primary cytoskeletal determinants of polarity in budding yeast. J. Cell Biol. 143, 1931-1945.
28. Evangelista, M., Pruyne, D., Amberg, D., Boone, C. & Bretscher, A. (2002). Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nature Cell Biology, 4, 32-41.
29. Pruyne, D., Evangelista, M., Yang, C., Bi., E., Zigmond, S., Bretscher, A. & Boone, C. (2002). Role of formins in actin assembly: nucleation and barbed end association. Science 297, 612-615.
30. Dong, Y., Pruyne, D. & Bretscher, A. (2003). Two Rho pathways converge to regulate formin-dependent actin assembly in yeast. J. Cell Biol. 161, 1081-1092.
31. Pruyne, D., Gao., L., Bi., E. & Bretscher A. (2004). Stable and dynamic axes of polarity utilize distinct forming isoforms in budding yeast. Mol. Biol. Cell, 15, 4971-4989.
32. Gao, L., Bretscher, A. (2008). Analysis of Unregulated Formin Activity Reveals How Yeast Can Balance F-Actin Assembly between Different Microfilament-based Organizations. Mol. Biol. Cell, 19, 1474-84.
33. Gao, L., Liu, W. & Bretscher, A. (2010). The yeast formin Bnr1p has two localization regions that show spatially and temporally distinct association with septin structures. Mol. Biol. Cell, 21, 1253-1262.
34. Liu, W., Santiago-Tirado, F. H. & Bretscher, A. (2012). Yeast formin Bni1p has multiple localization regions that function in polarized growth and spindle orientation. Mol. Biol. Cell, 23, 412-422.
35. Schott, D., Ho, J., Pruyne, D. & Bretscher, A. (1999). The carboxyl-terminal domain of a yeast myosin V has a direct role in secretory vesicle targeting. J. Cell Biol. 147, 791-807.
36. Schott, D., Collins, R. N. & Bretscher, A. (2002). Secretory vesicle transport velocity in living cells depends on the myosin-V lever-arm length. J. Cell Biol. 156, 35-39.
37. Yin, H., Pruyne, D., Huffaker, T. & Bretscher, A. (2000). Myosin V orientates the mitotic spindle in yeast. Nature 406, 1013-1015.
38. Santiago-Tirado, F. H., Legesse-Miller, A., Schott, D. & Bretscher, A. (2011). PI4P and Rab inputs collaborate in myosin-V-dependent transport of secretory compartments in yeast. Developmental Cell,20, 47-59.
39. Santiago-Tirado, F. H. & Bretscher, A. (2011). Membrane-trafficking sorting hubs: cooperation between PI4P and small GTPases at the trans-Golgi Network. Trends in Cell Biology, 21: 515-525.
40. Donovan, K. & Bretscher, A. (2012) Myosin-V is activated by binding secretory cargo and released in coordination with Rab/exocyst function. Developmental Cell, 23, 769-781.
41. Bretscher, A. (2003). Polarized growth and organelle segregation in yeast: the tracks, motors and receptors. J. Cell Biol. 160, 811-816.